As I have worked on various projects at Etsy, I have accumulated a suite of functions that help me quickly produce tables and charts that I find useful. Because of the nature of iterative development, it often happens that I reuse the functions many times, mostly through the shameful method of copying the functions into the project directory. I have been a fan of the idea of personal R packages for a while, but it always seemed like A Project That I Should Do Someday and someday never came. Until…
Etsy has an amazing week called “hack week” where we all get the opportunity to work on fun projects instead of our regular jobs. I sat down yesterday as part of Etsy’s hack week and decided “I am finally going to make that package I keep saying I am going to make.” It took me such little time that I was hit with that familiar feeling of the joy of optimization combined with the regret of past inefficiencies (joygret?). I wish I could go back in time and create the package the first moment I thought about it, and then use all the saved time to watch cat videos because that really would have been more productive.
This tutorial is not about making a beautiful, perfect R package. This tutorial is about creating a bare-minimum R package so that you don’t have to keep thinking to yourself, “I really should just make an R package with these functions so I don’t have to keep copy/pasting them like a goddamn luddite.” Seriously, it doesn’t have to be about sharing your code (although that is an added benefit!). It is about saving yourself time. (n.b. this is my attitude about all reproducibility.)
(For more details, I recommend this chapter in Hadley Wickham’s Advanced R Programming book.)
Step 0: Packages you will need
The packages you will need to create a package are devtools and roxygen2. I am having you download the development version of the roxygen2 package.
install.packages("devtools")
library("devtools")
devtools::install_github("klutometis/roxygen")
library(roxygen2)
Step 1: Create your package directory
You are going to create a directory with the bare minimum folders of R packages. I am going to make a cat-themed package as an illustration.
setwd("parent_directory")
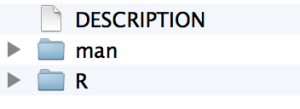
create("cats")
If you look in your parent directory, you will now have a folder called cats, and in it you will have two folders and one file called DESCRIPTION.
You should edit the DESCRIPTION file to include all of your contact information, etc.
Step 2: Add functions
If you’re reading this, you probably have functions that you’ve been meaning to create a package for. Copy those into your R folder. If you don’t, may I suggest something along the lines of:
cat_function <- function(love=TRUE){
if(love==TRUE){
print("I love cats!")
}
else {
print("I am not a cool person.")
}
}
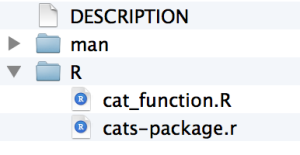
Save this as a cat_function.R to your R directory.
(cats-package.r is auto-generated when you create the package.)
Step 3: Add documentation
This always seemed like the most intimidating step to me. I’m here to tell you — it’s super quick. The package roxygen2 that makes everything amazing and simple. The way it works is that you add special comments to the beginning of each function, that will later be compiled into the correct format for package documentation. The details can be found in the roxygen2 documentation — I will just provide an example for our cat function.
The comments you need to add at the beginning of the cat function are, for example, as follows:
#' A Cat Function
#'
#' This function allows you to express your love of cats.
#' @param love Do you love cats? Defaults to TRUE.
#' @keywords cats
#' @export
#' @examples
#' cat_function()
cat_function <- function(love=TRUE){
if(love==TRUE){
print("I love cats!")
}
else {
print("I am not a cool person.")
}
}
I’m personally a fan of creating a new file for each function, but if you’d rather you can simply create new functions sequentially in one file — just make sure to add the documentation comments before each function.
Step 4: Process your documentation
Now you need to create the documentation from your annotations earlier. You’ve already done the “hard” work in Step 3. Step 4 is as easy doing this:
setwd("./cats")
document()
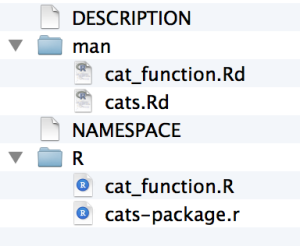
This automatically adds in the .Rd files to the man directory, and adds a NAMESPACE file to the main directory. You can read up more about these, but in terms of steps you need to take, you really don’t have to do anything further.
(Yes I know my icons are inconsistent. Yes I tried to fix that.)
Step 5: Install!
Now it is as simple as installing the package! You need to run this from the parent working directory that contains the cats folder.
setwd("..")
install("cats")
Now you have a real, live, functioning R package. For example, try typing ?cat_function. You should see the standard help page pop up!
(Bonus) Step 6: Make the package a GitHub repo
This isn’t a post about learning to use git and GitHub — for that I recommend Karl Broman’s Git/GitHub Guide. The benefit, however, to putting your package onto GitHub is that you can use the devtools install_github() function to install your new package directly from the GitHub page.
install_github('cats','github_username')
Step 7-infinity: Iterate
This is where the benefit of having the package pulled together really helps. You can flesh out the documentation as you use and share the package. You can add new functions the moment you write them, rather than waiting to see if you’ll reuse them. You can divide up the functions into new packages. The possibilities are endless!
Additional pontifications: If I have learned anything from my (amazing and eye-opening) first year at Etsy, it’s that the best products are built in small steps, not by waiting for a perfect final product to be created. This concept is called the minimum viable product — it’s best to get a project started and improve it through iteration. R packages can seem like a big, intimidating feat, and they really shouldn’t be. The minimum viable R package is a package with just one function!
Additional side-notes: I learned basically all of these tricks at the rOpenSci hackathon. My academic sister Alyssa wrote a blog post describing how great it was. Hadley Wickham gets full credit for envisioning that R packages should be the easiest way to share code, and making functions/resources that make it so easy to do so.

Thanks, that was very useful – I tried to do this years ago using the official R docuemntation, and quickly abandoned because it seemed too complicated for the use I had, using devtools, it seems super easy!
Absolutely useful!! The official documentation and even hadley’s is too “deep” for what I want to do.
Question: how would you save data?
Use the save() function from base R to save as a binary object loadable with load(), or write.table() to write data to a plaintext file.
Here’s a short article about including datasets in R packages; I found it helpful.
No, _here’s_ that article: http://kbroman.org/pkg_primer/pages/data.html
If you meant how do you include saved datasets so that they’re available after you install the package, make sure your data is present in your environment the form that you want with the name that you want, then call devtools::use_data() (filling in the appropriate arguments for your data). This will generate a /data directory under your parent directory, and place your .RData files there. After you rebuild the package the data should be available through the data() function, or however you normally prefer to load data from installed packages. For more detailed info, see http://r-pkgs.had.co.nz/data.html
Great post! Thanks a lot ! Cheers!
WOW! excellent post! thank you so much for sharing!
really Thanks for your post
and nice picture
<—
Thank you, it’s really helpful to start packaging quickly 😉
However if you feel intimidated to make a package, you can start by writing down your functions in an `.Rprofile` file.
That’s true! Although that can make it hard for reproducibility, since you don’t always think to share your .Rprofile when you share code. But that being said, better to start somewhere!
Thanks for sharing this. This is really great.
This is great. With the help of this post, I’ve just successfully made my own package and I’ve started using github as well.
Hi! Thank you very much!!!
This post were extremely helpful & instructive (knowing what is GitHub). However, I noticed, from the icons, that you are using a mac. Using a windows computer, I tried following the same instructions, but the .r package couldn’t be created. Or may be it was due to some other reason. Luckily, at the end, I managed to create the package, through rstudio. Mixed what you instructed and someone else’s instructions.
I wrote my first R package this weekend (convenience functions for geomorph using lab-standard settings) and your instructions made it so easy! Thanks.
thanks Hilary! This was very helpful!! Cheers from your old classmate and fellow hurdler 😉
Ahhh how cool! Glad you stumbled upon it. Chirp chirp!
Hello.
I’ve successfully created a simple package.
The package contains one function.
I’d like to know how to automatically execute that function whenever the package is loaded? (instead of waiting for the user to call it.
I learned how to do packaging with your tutorial. Thanks!
Reblogged this on The Falsifiable and commented:
This is so simple and exciting! I’ve written many R functions and never documented and recycled them. Think about all the time that I could have saved!
Watch out the format of comments you provide in the R file. It is quite tricky and confusing if you don’t read the guide on how to write description. It took me few hours to spot it and learn how to fix it. Pls read this article and don’t go through all the pain.https://cran.r-project.org/doc/manuals/r-release/R-exts.html#The-DESCRIPTION-file
Did I miss the part where you actually create the package?
Very helpful. A couple of questions. 1) Where/how do you document the package rather than individual functions. Is this in the description file? How do you modify it? 2) Most function documentation has a “Details” section. how do we add that?
I was wondering if anybody knows what Latex style to use to write a package’s reference manual. Thanks.
For windows
install.packages(“stringi”)
install.packages(“devtools”)
library(“devtools”)
devtools::install_github(“klutometis/roxygen”)
library(roxygen2)
Excellent Work!!!
By the way, for windows, one may need to do the following.
install.packages(“stringi”)
install.packages(“devtools”)
library(“devtools”)
devtools::install_github(“klutometis/roxygen”)
library(roxygen2)
Thanks! Bookmarked for future reference.
Thanks for posting this up – helped me through the process!
Hello and thanks for this primer on starting an R package 🙂 In order to gather candidate code to include, I was thinking about how one might find (almost) duplicate functions across all local .R files? After a few years of writing regular scripts for many different projects, they are scattered all across the harddrive for me.
You could call your package ‘whiskr’. Or maybe ‘mousr’. Thanks, that was super-helpful.
Hi Hillary,
Thanks for this! Definitely wonderful. Not to treat the comments section like a bug board, but maybe you’d have an idea on this:
When creating the package, my ‘cats-package.r’ file isn’t appearing. Thus, after installing I have documentation on commands (?etc works great), but nothing is actually loaded. It makes a great glossary, but not very functional. Does something like this seem familiar, or simple?
This is just so awesome and helpful. FYI roxygen2 is now on CRAN